Design
Avogadro started with a single idea. All great editors are improved by plugins.
Everyone accepts the idea that Photoshop or GIMP or Eclipse have plugin modules which add features, change the interface, etc. These programs do not attempt to solve image or text editing for everyone. Instead, they provide some core functionality to be extended by others.
This is even more important in the area of chemistry, where many fields come together and different users have vastly different needs. Biochemists need protein ribbon diagrams and residue builders. Crystallographers need unit cells and crystal face renderings. The list goes on.
The original design outline was written by Geoff Hutchison.
Goals
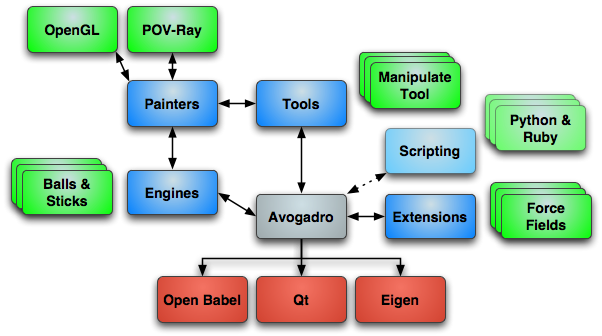
The project has several key goals, including being easy to use, quickly responsive, flexible and extensible. Speed of response indicates that many elements of the program should be multi-threaded and optimized for speed. Flexibility and extensibility are reflected in the plugin architecture described below.
Essentially, the program should scale – both in terms of speed (usable on old hardware, fast on multi-core systems), and in terms of functionality.
Further Design
Avogadro is intended to be cross-platform (Mac, Windows, Linux), so use of OpenGL graphics and the Qt toolkit are used. As an open source project, others can improve the code, both the core and the plugin modules. Reuse of existing frameworks such as Open Babel and Kalzium is recommended.
The interface must be user-friendly for a general audience: from students to experts.
The plugin APIs for rendering, mouse tools, commands, etc. need to be well-written, stable, and well documented. This should also allow access from multiple programming environments: scripting languages (Perl, Python, Ruby, shell script), Java, C++, etc.
The framework should allow features to be added and removed to customize features (e.g., for a student course)
The program should be international – localization must be possible.
A flexible rendering system should allow “overlays” of multiple rendering engines. For example, some protein views show a ribbon for the amino acid backbone, as well as wireframe of all atoms. Labels, surfaces, colors should all be customized.
Rendering should allow for specific atoms, bonds, residues, etc. to be sent to a custom engine. For example, to highlight specific atoms, the radius might be larger (e.g., the full van der Waals radius).
Engines should allow for animation, crystal cell repeats, etc.
Custom primitives should eventually be possible:
- Atoms, bonds, residues, etc.
- Protein and nucleic ribbons and cartoons
- Polyhedra for crystals
- Polymer chains
- Surfaces and volumes
- Slices (e.g., plane through a specific surface or volume)
- Arrows or lines for notation
- Custom text labels
Exporting graphics should be easy:
- Standard bitmap formats (PNG, GIF, JPEG, TIFF)
- PostScript and vector formats (PS, EPS, SVG, PDF)
- External render methods (PovRay)
- Animation / movie formats
Mouse action should be extensible. Tools should include standard elements:
- View the molecule and move the camera (translate, rotate, zoom, etc.)
- Draw and erase atoms and bonds
- Move and translate atoms and fragments
- Select by clicking and dragging
- Measure charges, distances, angles, and dihedral angles
All of these should be extensible via a tool API, not just by changing the code of existing tools, but also by adding new plugins which operate in the Avogadro framework.
Standard user interface conventions should be supported, including undo/redo, cut/copy/paste, etc.
Commands for both quick actions and long computation should be possible. An “extension” API should be available to support:
- Manipulating the molecular model
- Changing rendering options
- Selecting atoms and other objects
- Access to external programs
- Scripting via Python, Perl, Ruby, etc.